Week 9 has become the week dedicated to my analysis of my project. As this semester draws to a close and my projects data becomes clear how to interpret my data and explain my results. To begin my experiment I identified the bacteria which was given to me as an unknown, through a series of tests and a couple inconclusive tests I identified Bacillus subtilis was my unknown bacteria. Knowing the bacteria which I was working with I proceeded to perform a DNA extraction of my bacteria using 2 kits, from these 2 kits I would be able to compare quantity vs quality of the DNA. I reviewed the DNA extraction protocol for the E.Z.N.A. Bacterial DNA kit, but prior to beginning the DNA extraction I was prompted to a similar experiment that would yield results for a better purpose. My experiment was slightly altered, from comparing the DNA extractions of 2 kits to comparing the DNA extraction of 1 kit to the DNA extraction using egg white lysozymes. I would now test the DNA kit and egg white on my new day 1 of the experiment to set a bench mark or baseline for DNA extraction yields. Followed by a DNA extraction using the egg white every week after to gather data on the shelf life of the egg white lysozyme.
Thursday, December 8, 2016
Tuesday, December 6, 2016
Week 10
The research phase of my experiment is officially over, as I analyze the data I come to realize egg white is not viable for a long time. To be exact egg white lysozyme is of no use after 1 week. The sample from Bacillus subtilis has come out a yield rate of less than half. This week has been dedicated to my research paper and formatting the data properly so others can understand the protocol I used. Without being able to fully explain my process and having someone reattempt my experiment I cannot prove a successful experiment. I believe the most important thing to learn here is not the actual experiment but the process of a science experiment: trial and error, questioning steps and how they work, but most important of all to me would be the ability to hand some one else my notebook and they can use what i have done not only as an example but a guide to conduct the experiment. I have to admit my notebook this semester is not the cleanest, nor is it detailed enough for someone to repeat my experiment. In time I plan on taking this mistake and all other mistakes I have made throughout this semester and finding how I can do what I did wrong better next time.
Thursday, November 17, 2016
Week 8
My nano-drop tests for this week have really affected my experiment. My hypothesis was right, the shelf life of lysosomes from egg whites for DNA extractions is not very long, less then 1 week to be exact. After last weeks DNA extraction and nano-drop, i repeated the exact same process this week only to obtain half the DNA results from 1 week ago. This brings the question how can I alter the lysosomes to extend its shelf life without compromising its chemical properties that allow for it to be used in DNA extraction? Can I partially prep the egg white and would the shelf life be greater? or to be more specific should I test a DNA extraction everyday to know exactly how long the shelf life really is? For now I will draft up my research paper upon this experiment drawing my final conclusions about the experiment and my obstacles and mistakes throughout this wonderful learning experience. This is a picture of my original Bacteria testing before getting into the DNA extractions.
Thursday, November 10, 2016
Week 7 Back in the Lab
So after a successful robotics competition I am happy to be back in the lab working with the extraction of lysozyme from egg white. Due to having my previous lysozyme extraction sit unused for too long I dedicated this week to create a fresh batch of lysozyme and a test of DNA extraction from the fresh batch. After the batch was freshly made I completed a DNA extraction from my original unknown bacteria using a DNA extraction kit and my lysozyme in order to compare and create a baseline of data for the following weeks. Using a nano-drop technique I tested the DNA extractions for purity and successful extraction.
Thursday, November 3, 2016
Week 6 Vex competiton
Due to a upcoming competition, this week I have sidelined my time in the lab in order to complete and finalize my plan for my robot in order to compete this weekend. From the beginning till now the robot has been taken apart and put together several times and each time being a different robot performing the same task. Now I have finalized my design choice for this competition with a scorpion style design. sacrificing speed for torque and the ability to raise a cube over the fence, still some work is in progress with 2 days left. My center of mass for the robot needs adjusting due to the weight of a cube tilting the robot over rendering it useless. I'm not sure if I should add a wheelie bar style assistance to it or adjust the center of mass over the rear tires to allow the cube to be over the center of the chassis. That is my goal for tomorrow experiment with both options and make a final decision for the competition.
Thursday, October 27, 2016
Week 5 DNA Extraction
After a reading up on the process of using the Omega DNA extraction kit I was shown a much faster yet not as efficient process to obtain the same results. Today I took lysozyme I have purified and run a DNA extraction side by side with the extraction process given to me. My process consists of using my unknown bacteria as the subject to which my DNA extraction will occur. The three DNA extraction tests I ran today were the use of a lysozyme I extracted from egg whites, lysome provided in a kit, and 40% ethanol in order to compare the results and effectiveness of each.
Thursday, October 20, 2016
Week 4 Lysozyme Extraction
Egg
Lysosome in DNA Extraction
The topic
is significant because of Lysozyme’s ability to break chemical bonds on outer
walls of bacteria. It is important to learn more about lysozymes to understand
why and how they break down chemical bonds on outer walls of bacteria. It is
important to conduct research in this area so we can find ways to apply
lysozymes ability to break down chemical bonds in other scientific fields.
·
Experiment- I will run
a procedure of partial purification of lysozyme.
1. For the partial purification of lysozyme, egg
whites, carefully separated from the egg yolks, were diluted 3- or 3.3-fold
with 0.05M NaCl solution.
2. To precipitate the egg white proteins other
than lysozyme, the pH of this mixture was set to 4.0 by carefully adding
several drops of 1 N acetic acid and it was diluted with an equal volume of
40%(v/v) ethanol. After 30-minute incubation at room temperature in the
presence of ethanol, the mixture is centrifuged at 15,000xg for 15 minutes at 4
degrees C: then the precipitates were discarded.
3. The supernatants were analyzed for their
lysozyme activity and protein content to determine if lysozyme extraction was
successful.
4. A
DNA extraction is performed using a kit the epicenter QuickExtract Bacterial
DNA Extraction Kit (QEB0905T) and E.Z.N.A. Bacterial DNA Kit from Omega bio-tek
(D3350-00) as controls
5. A
DNA extraction is performed using both kits but replacing the kit provided
lysozyme with the student prepared egg white (EW) lysozyme.
|
Name
|
I/D/C
|
Units
|
Comment
|
|
Eggs
|
C
|
-
|
-
|
|
Lysozyme
|
D
|
-
|
-
|
|
Partial Purification of Lysozyme Protocol
|
I
|
|
Refer to Protocol
|
Is it possible to extract lysozyme from egg whites?
My hypothesis is that the extraction of lysozymes from egg whites will be
possible. The results I may discover are that the purity of the lysozyme from
egg white may not be as pure as we need for a DNA extraction.
References
·
Alderton, G., Ward, W.H.,
& Fevold, H.L. (1945). Isolation of Lysozyme From Egg White [PDF]. Journal of Biological Chemistry, (157),
43-58. Retrieved from http://www.jbc.org/content/157/1/43.full.pdf
·
Fakharany, E. E., &
Hassan, M. (2016). A universal method for extraction of genomic DNA from
various microorganisms using lysozyme. New
Biotechnology, 33(1), S210.
http://dx.doi.org/10.1016/j.nbt.2016.06.1445
·
Ganong, B. (2007, June 11).
Isolation of Lysozyme from Egg White. Retrieved October 20, 2016, from Barry
Ganong's Homepage website:
http://faculty.mansfield.edu/bganong/biochemistry/lysozyme.htm
·
QuickExtract™ bacterial DNA extraction kit [pdf]. (2012, December). Retrieved from
http://www.epibio.com/docs/default-source/protocols/quickextract-bacterial-dna-extraction-kit.pdf?sfvrsn=8
·
Xue-li, C., Ting, L., &
Ito, Y. (2007). Separation of Chicken Egg-White Lysozyme by High-Speed
Countercurrent Chromatography using a Reverse Micellar System. Journal of Liquid Chromatography &
Related Technologies, 30(17),
2593-2603. http://dx.doi.org/10.1080/10826070701540555
Friday, October 14, 2016
Week 3 Unknown Bacteria
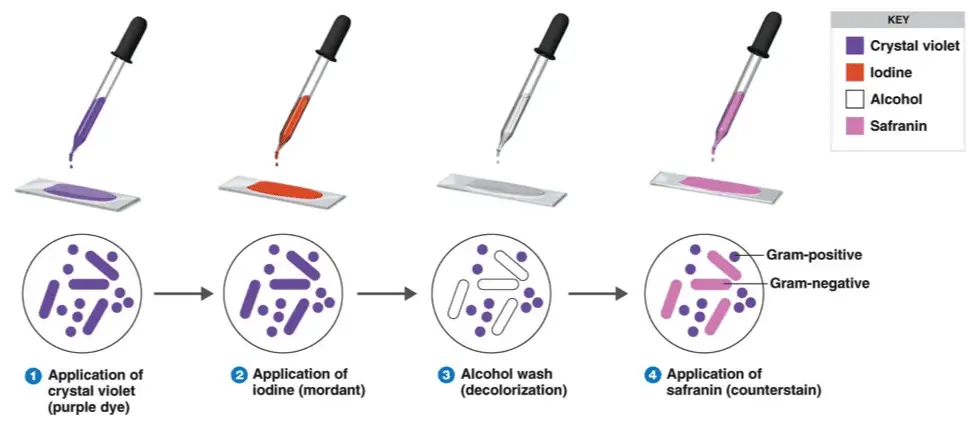
Today I look forward to receiving my sample of unknown bacteria and attempting to identify it using the techniques previously discussed, including but not limited to Gram Staining and Ziel-Neelsen Staining. If the results prove to be effective i will proceed with the results to the next step, but if the tests are ineffective i will research a third way to which i can attempt to identify the bacteria.
After receiving my sample of unknown bacteria I have begun my identification process by using a three quadrant streak plate method. I took a sample of the unknown and streaked 1/3 of the TSA petri-dish. I took a second sample of the same unknown bacteria and streaked a second time beginning in the prior quadrant for one drag of the loop with the bacteria and streaking another 1/3 of the petri-dish, finalized by a third sample of the same unknown. Dragging it once through the 2nd quadrant and continuing to streak the 3rd and final quadrant. I then placed the petri- dish in the incubator which will allow for the bacteria to colonize over the next 24 hours.
After receiving my sample of unknown bacteria I have begun my identification process by using a three quadrant streak plate method. I took a sample of the unknown and streaked 1/3 of the TSA petri-dish. I took a second sample of the same unknown bacteria and streaked a second time beginning in the prior quadrant for one drag of the loop with the bacteria and streaking another 1/3 of the petri-dish, finalized by a third sample of the same unknown. Dragging it once through the 2nd quadrant and continuing to streak the 3rd and final quadrant. I then placed the petri- dish in the incubator which will allow for the bacteria to colonize over the next 24 hours.
Thursday, October 6, 2016
Week 2 Unknown Bacteria
To begin my first day here at the S-STEM internship I will
begin research in regards to how to identify bacteria. From my knowledge of
bacteria I know Gram Staining can be an effective form of identifying Gram
Positive and Gram Negative bacteria. Also I understand that some bacteria have
a "wax" like outer layer that would prove Gram Staining to be
ineffective in the process of identifying the bacteria type. Therefore when
Gram Staining proves ineffective another process of identification can be
Ziehl-Neelsen Staining. This staining process proves effective only when
identifying Mycobacteria in general. Ziehl-Neelsen Staining cannot provide
specific identification of species within Mycobacteria, but can prove that the bacteria
being tested are Mycobacteria. I understand the general concepts of the
staining processes but have never attempted them nor do I know the specific
process in which they are performed. My goal is to research the specific
process for these staining methods and attempting them on unknown bacteria to
better understand how the staining looks when performed. Once I have completed
this, my following goal would be to research a way to differentiate between 2
different types of a bacteria genus. I also look forward to sharing my results
and comparing with other interns our processes and findings in order to
understand maybe other ways I can accomplish the same task or a more effective
way or obtaining the same result. Also I would like to find a way to identify
the sample without compromising it so I can conduct further testing with one
sample.
Subscribe to:
Comments (Atom)